Overview of Major Features
If you already own an older copy of MacVector, and would like to know what new features have been added since the version you own, you can find that information on this summary page.
Another piece of brilliant software from Mekentosj and a bargain at only 29 euros ($40). Geneious Geneious is a software package of genome & proteome research tools for protein, DNA, or molecular visualization, literature searching and storageand more. The Pro version does LOTS more. DNA Master is a freeware. DNA sequence editor and analysis package. I only ask that users register the program and send interested parties here to pick up a version. Thank you for downloading DNA Master from our software library. The download version of DNA Master is 5.22.1. The software is periodically scanned by our antivirus system. There are many applications for opening DNA sequence files on the PC, but few exist for opening them on the Mac. Furthermore, none of the existing software will generate quality reports for your DNA sequence chromatograms. To use this DNA sequence analysis software, simply drag and drop chromatogram files onto the program's icon. Sequence / Chromatogram Viewing Software A number of free software programs are available for viewing trace or chromatogram files. Click on the appropriate icon(s) to go to the respective Web page. The Mac Sequence View Software suite comes with three programs to enhance your productivity when performing DNA sequence analysis. The main program - Mac Sequence View - is a drag and drop.
Graphical Sequence Editing
MacVector can read and write DNA and Protein sequences in most popular file formats. In addition to directly editing sequences and features/annotations, MacVector has an intuitive 'Click Cloning' graphical interface that lets you easily replicate laboratory cloning experiments to create new molecules. MacVector uses the native Mac OS X Quartz graphics to generate publication quality images that can be scaled to any size with no loss of resolution. More details...
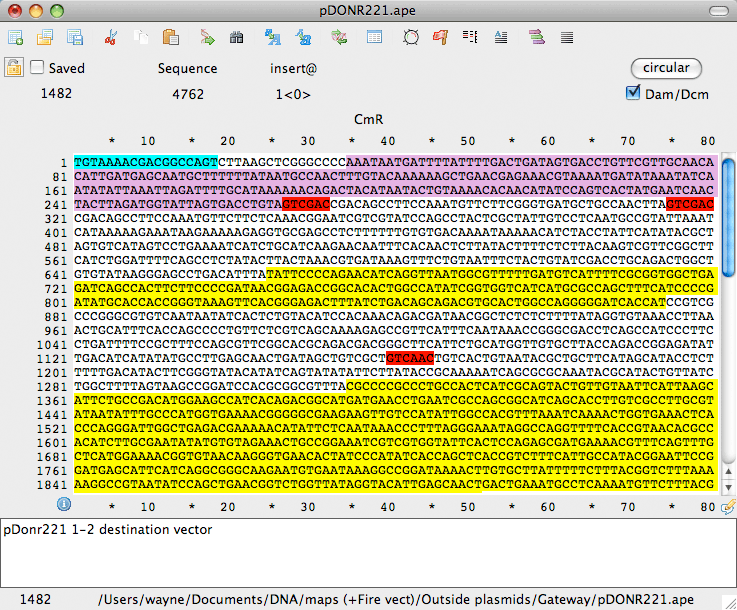
Gateway and Topo Cloning
MacVector supports the popular Gateway, TOPO TA and Zero Blunt cloning technologies from Invitrogen. With a few simple clicks from an intuitive graphical interface, you can replicate your biological manipulations at the bench to create new molecules with the correct sequences across the cloning and recombination junctions. More details...
Auto Annotation
You can scan an unannotated or partially annotated sequence against a folder on your hard drive and MacVector will identify matching features in sequences in the target folder and add them to your sequence. Because MacVector includes custom feature appearance information when annotating the sequence, you can use this to maintain a carefully curated set of your favorite genes and sequences each with a graphical appearance that best suits your needs. More details...
Primer Design
You can design primers for either PCR or Sequencing/Hybridization probes using a variety of primer design functions, including an interactive manual design interface along with several scanning functions such as the popular Primer3 algorithm. The interactive interface clearly shows which potential primers may have secondary structure problems, alternate binding sites or other characteristics that might impact their use in experiments. More details...
DNA Analysis
MacVector provides a wide variety of useful DNA analysis tools, including base composition analysis, Restriction Enzyme searches, DNA Subsequence searches and 'Dot-Plot' comparisons between DNA:DNA and DNA:Protein sequences. A Coding Preference toolbox lets you select a variety of algorithms to graphically scan a DNA sequence for likely protein coding open reading frames. More details...
Protein Analysis
Protein sequences can be reverse translated into DNA, compared using 'Dot-Plot' analysis and scanned for Proteolytic cleavage sites and amino acid sequence motifs. A comprehensive Protein Analysis Toolbox provides a wide variety of algorithms for analyzing the composition of proteins and presenting the results in graphical and tabular formats. More details...
Database Searching
MacVector has built-in Internet connectivity to the NCBI BLAST and Entrez databases. You can directly search Entrez for DNA or Protein sequences based on features, authors, keywords etc and directly download them into MacVector, complete with all features and annotations. The built-in BLAST interface lets you submit multiple BLAST jobs using DNA or Protein sequences and then download any matching sequences by selecting them from a hit list. Even without an Internet connection, MacVector can align sequences against any folder on your hard drive using a FastA algorithm, allowing for 'local' database searches. More details...
Multiple Sequence Alignment
You can align unlimited numbers of DNA or Protein sequences using the ClustalW algorithm built in to MacVector. A full-featured editor lets you make manual adjustments to the alignments and view them using a wide variety of customizable color schemes. You can create publication quality graphical outputs of the alignments and view pairwise combinations of the sequences in aligned and matrix formats. More details...
Sequence Assembly
Sequence Assembly functionality in MacVector is now provided by two modules. First there is a built-in function called Align To Reference. This can be used for two different functions, Sequence Confirmation and cDNA Alignment. This is included with MacVector and allows you to import trace files or sequence files and assemble them against a template sequence. This is ideal for small scale sequencing projects, especially resequencing. For example, checking an in situ mutagenesis experiment, a construct you've just made, or confirming the sequence of a cloned PCR fragment. It's also an excellent tool for SNP analysis, with some special tools to allow you to easily spot mutations from your original template sequence.
For full scale sequencing projects where you do not know the sequence you'll need MacVector and a separately purchased plugin called Assembler. This is a full contig assembly application that uses the phred, phrap and cross_match algorithms from the University of Washington to assembles traces into contigs. It displays full quality scores of the reads and the aligned contigs. The trial version of MacVector also includes the Assembler plugin. Click here to go to the Assembler product page.
Please note that AssemblyLIGN is no longer included with MacVector.
Chromatogram Trace Viewers
These programs are limited in scope but serve a good purpose in evaluating the quality of data produced by allowing users to view the raw data associated with .ab1 files.
Protein Sequence Analysis Software
FinchTV (Windows/OSX)
From the makers of our data management system. Displays quality scores and much more. We really like the feature that links directly to NCBI for a BLAST search! What is more the application is FREE!
4Peaks (OSX Only)
Written to run natively on OSX and makes full use of the OSX interface. The application has lots of features and is more than just a “chromatogram viewer”. The best feature is the application is totally FREE! The same people have other OSX applications that might be useful for scientists.
ABI Seq Scape(Windows)
Another free viewer. This program is provided by the maker of the machine we run your samples on.
Chromas LITE (Windows Only)
This application will allow PC users to display, edit and print Applied Biosystems chromatogram files. You can even translate the sequence and reverse complement it! There is a ‘Pro’ version with more features.
Traceviewer(Windows/OSX/Linux)
This is a Java program that allows the reading of sequence traces from ABI or SCF files, the display of traces on white or black background and the display of Phred quality values as bars or numbers with one of three different color schemes. TraceViewer is free for academic users. Traceviewer for the Macintosh does not suffer from the same problem as Editview.
Contig Assembly and Sequence Alignment
These software packages can be used to view, edit, assemble, align, BLAST and even design primers. Some of the programs have phylogenetic analysis software built in as well! For more information, click on the links below.
Proprietary Software:
Proprietary software may cost more, but it also tends to be more current and offers long term and ongoing support.
- CLCbio DNA Workbench (Windows/OSX/Linux) Available for use on workstations in our facility
- Bioinformatics Toolbox 3.1 for MATLAB (Windows/OSX/Linux)
- CodonCode Aligner (Windows/OSX)
- Geneious(Windows/OSX/Linux)
- iNquiry (OSX Server/Unix/Linux)
- Lasergene (Windows/OSX)
- Sequencher (Windows/OSX) v4.2 available on Mac workstations in our facility
Free and Open Source Software:
Free, customizable and often times you can examine the source code. These can also be difficult to find support for.
- BioEdit (Windows only)
- EMBOSS (OSX/Unix/Linux)
- MEGA4 (Windows/OSX/Linux)
- Gene Construction Kit (Windows/OSX)
- MacVector (OSX only)
- Peaks (Windows/OSX/Linux)
- Unipro UGENE (Windows/OSX/Linux)
Analysis Software geared toward the interests of Evolutionary Analysis, Population Genetics, Phylogenetics and the like:
A List of Phylogeny Programs by Joe Felsenstein If you can’t find what you’re looking for in this comprehensive and exhaustive list, it probably doesn’t exist.
JMATING a software for the analysis of sexual selection and sexual isolation effects from mating frequency data.
Mesquite is software for evolutionary biology, designed to help biologists analyze comparative data about organisms. Its emphasis is on phylogenetic analysis, but some of its modules concern population genetics, while others do non-phylogenetic multivariate analysis.
TREEFINDER is a software to compute phylogenetic trees from molecular sequences.
TreeRot is a very easy program to use for determining Bremer indices (decay indices) for phylogenetic trees. It is also capable of providing partitioned DI, and after your analysis is complete, the program can parse your output log and give you the DI at each node (so you don’t have to go through the log file yourself and do the arithmetic).
SGRunner SG Runner is a graphical user interface for Seq-Gen.
Cadence Cadence helps to conduct Bayesian relative rates tests.
PolypSizer A program for measuring things. Very useful when combined with a digitizing pad and drawing tube.
Gel Reader A program for estimating band sizes on electrophoretic gels.
Dna Sequence Editing Software
For Windows:
Acquity 4.0 (ccg12 and ccg13) | MrBayes 3.0b4 |
Clustalx 1.83 | PAUP 4.0 |
DnaSP 4.0 | Phylip 3.65 |
GenePix Pro 6.0 (ccg12 and ccg13) | Phyml 2.4.4 |
Genespring 7.2 (ccg12 and ccg13) | Seq Scape 2.1.1 |
Mega 2 | Sequencing Analysis 5.1.1 |
Mega 3.1 | TreeView |
For Macs:
Clustalx 1.83.1 |
MacClade 4.06 |
MrBayes 3.1.2 |
PAUP 4.0 |
Phylip 3.65 |
Phyml 2.4.4 |
TreeView 1.6.6 |
CoMET (Continuous-character Model Evaluation and Testing) is a Mesquite module for calculating the likelihood of different Brownian motion-based models of continuous character change on phylogenies
Modeltest A tool to select the best-fit model of nucleotide substitution
MrAIC Perl script for selecting DNA substitution models using PHYML
MrModeltest C program for selecting DNA substitution models using PAUP Mailfit: Perl script that runs PAUP and MrModeltest2 (or Modeltest) and sends (optionally) results via e-mail
Burntrees Perl script for manipulating MrBayes tree and parameter files. The script comes with an extra tool, catmb.pl, for concatenating files from separate MrBayes runs.
Vim Nexus syntax highlighting Nexus syntax highlighting files for the Vim editor. Recognizes many/most of the PAUP/MrBayes commands
Vim Nexus syntax highlighting Vim highlighting files for DNA sequence alignments
MrModelfit/Modelfit Perl script for running MrModeltest2/Modeltest
BootPHYML Perl script for performing non-parametric bootstrapping using PHYML